In the intricate dance between microbes and the human immune system, there exists a fascinating survival strategy employed by various microbes, including bacteria, parasites, and fungi. This strategy involves the cleavage of antibodies, specifically immunoglobulins, which are crucial components of the body’s defense mechanism.
The Impact of Chronic Stress on the Immune System
Before delving into the intricate mechanisms of microbial survival strategies, it’s essential to understand the context in which these interactions occur. Chronic stress and exhaustion can significantly compromise the immune system, rendering individuals more susceptible to infections and illnesses. This weakened state provides an opportune environment for microbes to exploit and thrive.
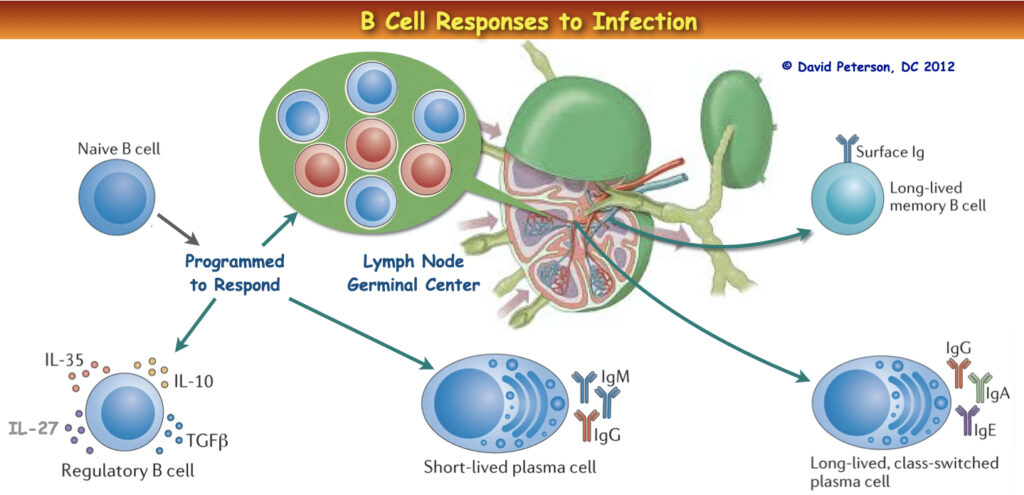
Secretory Immunoglobulin A (SIgA): Guardian of Mucosal Surfaces
At the forefront of mucosal defense stands Secretory Immunoglobulin A (SIgA), the principal immunoglobulin found in the mucosal secretions of the gastrointestinal tract. SIgA plays a vital role in protecting the gastrointestinal mucosal lining against inhaled and ingested pathogens. It acts as a first line of defense by inhibiting the colonization of potentially harmful microorganisms at these surfaces.
Microbial Survival Strategy: Cleaving Immunoglobulins
Microbes have evolved sophisticated mechanisms to circumvent the body’s immune defenses, with one notable strategy being the cleavage of immunoglobulins. By cleaving antibodies such as SIgA, microbes can undermine the effectiveness of the immune response, facilitating their survival and proliferation within the host.

Enzymatic Cleavage by Pathogenic Microbes
A plethora of pathogenic bacteria and parasites produce enzymes capable of cleaving SIgA into fragments, rendering it ineffective in combating microbial invasion. These enzymes target the structure of immunoglobulins, disrupting their function and compromising the host’s defense mechanisms.
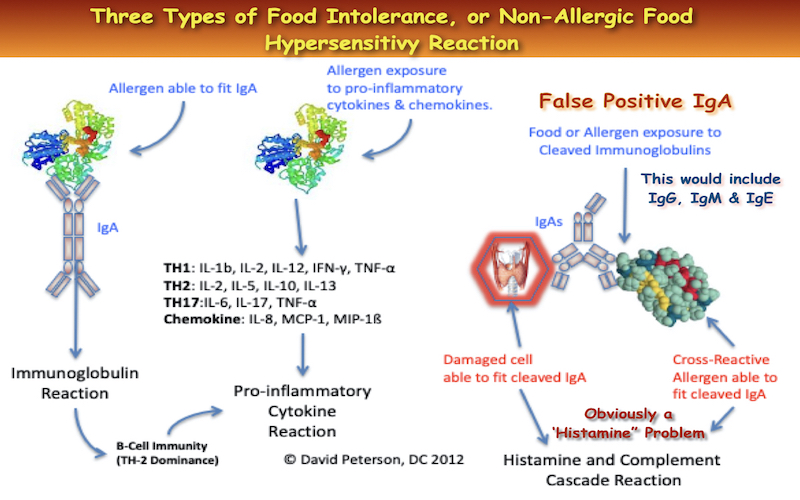
Microbial Diversity in Cleaving Immunoglobulins
The Genova GIFX, a rich ecosystem inhabited by diverse microbial species, harbors numerous microbes equipped with the ability to cleave not only SIgA but also other immunoglobulins such as SIgM, SIgG, and SIgE. Among these microbes are Bacteroides, Prevotella, Clostridia, Mycoplasma, Pseudomonas, fungi, and hookworm, each wielding enzymatic machinery tailored to cleave specific antibody types.
The immune system may become compromised due to chronic stress and exhaustion, making individuals more susceptible to infections and illnesses. Microbes (bacteria, parasites and fungi) employ a survival strategy of hacking into the immune system.
Secretory immunoglobulin A (SIgA) protects the gastrointestinal mucosal lining against inhaled and ingested pathogens. Secretory IgA (SIgA) is the principal immunoglobulin present in the mucosal secretions of the gastrointestinal tract. SIgA is thought to play a major role in defense at these surfaces by inhibiting the colonization of potentially pathogenic microorganisms. Microbes are capable of damaging immunoglobulins as part their survival mechanism. 1Qing Han, Elizabeth M. Bradshaw, Björn Nilsson, David A. Hafler, J. Christopher Love. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab on a Chip, 2010; 10 (11): 1391 DOI: 10.1039/b926849a

This microbial survival strategy includes:
- A number of bacteria that are mucosal pathogens of humans produce enzymes that cleaves the SIgA into fragments. Many pathogenic bacteria and parasites produce SIgA enzymes that cleave SIgA, making SIgA ineffective and testing questionable.2Koteswara R. Chintalacharuvu, Philip D. Chuang, Ashley Dragoman, et al. Cleavage of the Human Immunoglobulin A1 (IgA1) Hinge Region by IgA1 Proteases Requires Structures in the Fc region of IgA. Infect Immun. 2003 May; 71(5): 2563–2570. doi: 10.1128/IAI.71.5.2563-2570.2003 PMCID: PMC153282,3Fujiyama, Y., M. Iwaki, K. Hodohara, S. Hosoda, and K. Kobayashi. 1986. The site of cleavage in human alpha chains of IgA1 and IgA2: A2m(1) allotype paraprotein by the clostridial IgA protease. Mol. Immunol. 23:147–150.,4Fujiyama, Y., K. Kobayashi, S. Senda, Y. Benno, T. Bamba, and S. Hosoda. 1985. A novel IgA protease from Clostridium sp. capable of cleaving IgA1 and IgA2m(1) but not IgA2m(2) allotype paraproteins. J. Immunol. 134: 573–576.,5Anna Eriksson and Mari Norgren. Cleavage of Antigen-Bound Immunoglobulin G by SpeB Contributes to Streptococcal Persistence in Opsonizing Blood. Infect. Immun. 2003, 71(1):211. DOI: 10.1128/IAI.71.1.211-217.2003.,6Puthia MK, Vaithilingam A, Lu J, Tan KS.Degradation of human secretory immunoglobulin A by Blastocystis.Parasitol Res. 2005 Nov;97(5):386-9. Epub 2005 Sep 7.,7Lei B, DeLeo FR, Hoe NP et al. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat Med 2001; 7: 1298–1305.,8Garcia-Nieto RM, Rico-Mata R, Arias-Negrete S, Avila EE. Degradation of human secretory IgA1 and IgA2 by Entamoeba histolytica surface-associated proteolytic activity. Parasitol Int. 2008 Dec;57(4):417-23. Epub 2008 May 15.,9Xuchu Que and Sharon L. Reed. Cysteine Proteinases and the Pathogenesis of Amebiasis Clin Microbiol Rev. 2000 April; 13(2): 196–206. PMCID: PMC100150,10E V Frandsen, J Reinholdt, and M Kilian. Enzymatic and antigenic characterization of immunoglobulin A1 proteases from Bacteroides and Capnocytophaga spp. Infect Immun. 1987 March; 55(3): 631–638. PMCID: PMC260386
- Many of the microbes found in the Genova GIFX have the ability to cleave SIgA, SIgM, SIgG and SIgE, i.e. Bacteroides,11Gregory RL, Kim DE, Kindle JC, Hobbs LC, Lloyd DR.Immunoglobulin-degrading enzymes in localized juvenile periodontitis. J Periodontal Res. 1992 May;27(3):176-83. Prevotella,12Senior BW, Dunlop JI, Batten MR, Kilian M, Woof JM. Cleavage of a recombinant human immunoglobulin A2 (IgA2)-IgA1 hybrid antibody by certain bacterial IgA1 proteases. Infect Immun. 2000 Feb;68(2):463-9.,13Frandsen EV, Reinholdt J, Kjeldsen M, Kilian M. In vivo cleavage of immunoglobulin A1 by immunoglobulin A1 proteases from Prevotella and Capnocytophaga species. Oral Microbiol Immunol. 1995 Oct;10(5):291-6.Clostridia,14Qiu J, Brackee GP, Plaut AG. Analysis of the specificity of bacterial immunoglobulin A (IgA) proteases by a comparative study of ape serum IgAs as substrates. Infect Immun. 1996 Mar;64(3):933-7.,15Reinholdt J, Kilian M. Interference of IgA protease with the effect of secretory IgA on adherence of oral streptococci to saliva-coated hydroxyapatite. J Dent Res. 1987 Feb;66(2):492-7.Mycoplasma,16Cizelj I, Bercic RL, Dusanic D, Narat M, Kos J, Dovc P, Bencina D. Mycoplasma gallisepticum and Mycoplasma synoviae express a cysteine protease CysP, which can cleave chicken IgG into Fab and Fc. Microbiology. 2011 Feb;157(Pt 2):362-72. Epub 2010 Oct 21. Acinetobacter,17Morse SA. Neisseria, Moraxella, Kingella and Eikenella. In: Baron S, editor. SourceMedical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 14. Streptococcus,18Senior BW, Batten MR, Kilian M, Woof JM. Amino acid sequence requirements in the human IgA1 hinge for cleavage by streptococcal IgA1 proteases. Biochem Soc Trans. 2002 Aug;30(4):516-8.,19Hulting G, Flock M, Frykberg L, Lannergård J, Flock JI, Guss B. Two novel IgG endopeptidases of Streptococcus equi. FEMS Microbiol Lett. 2009 Sep;298(1):44-50. Pseudomonas,20Kharazmi A. Mechanisms involved in the evasion of the host defence by Pseudomonas aeruginosa. Immunol Lett. 1991 Oct;30(2):201-5. Fungus,21Silva BA, Pinto MR, Soares RM, Barreto-Bergter E, Santos AL. Pseudallescheria boydii releases metallopeptidases capable of cleaving several proteinaceous compounds. Res Microbiol. 2006 Jun;157(5):425-32. Epub 2006 Jan 20. Hookworm,22Loukas A. Prociv. Immune Responses in Hookworm Infections. Clin Microbiol Rev. 2001 October; 14(4): 689-703,23Pritchard DI. The survival strategies of hookworms. Parasitol Today. 1995;11:255-259 Schistosome.24McIntosh RS. Jones FM. Dunne DW. McKerrow JH. Pleass RJ. Characterization of immunoglobulin binding by schistosomes. Parasite Immunology 2006, 28, 407-419
- Bacteria cause lymphocyte death by enhancing the cell-death programing of immune cells. The presence of apoptotic lymphocytes down-regulates early innate immunity, creating a permissive environment for bacterial growth.25Faherty CS, Maurelli AT. Staying alive: bacterial inhibition of apoptosis during infection. Trends Microbiol. 2008;16(4):173-180. doi:10.1016/j.tim.2008.02.001,26JA. Carrero, ER. Unanue. Lymphocyte apoptosis as an immune subversion strategy of microbial pathogens. Trends in Immunology, Vol 27, ISSUE 11, P497-503, NOVEMBER 01, 2006, DOI:https://doi.org/10.1016/j.it.2006.09.005,27Hotchkiss RS, Tinsley KW, Swanson PE, et al. Prevention of lymphocyte cell death in sepsis improves survival in mice. Proc Natl Acad Sci U S A. 1999;96(25):14541-14546. doi:10.1073/pnas.96.25.14541
- Microbes have the ability to hack the immune system to evade or become invisible to White Blood Cells as part of their survival strategy.28Frandsen, E. V. G., J. Reinholdt, and M. Kilian. 1991. Immunoglobulin A1(IgA1) proteases from Prevotella (Bacteroides) and Capnocytophaga species in relation to periodontal diseases. J. Periodontal Res. 26:297–299.,29Frandsen, E. V. G., J. Reinholdt, M. Kjeldsen, and M. Kilian. 1995. In vivo cleavage of immunoglobulin A1 by immunoglobulin A1 proteases from Prevotella and Capnocytophaga species. Oral Microbiol. Immunol. 10:291–296.,30Frandsen, E. V. G., J. Reinholdt, and M. Kilian. 1987. Enzymatic and antigenic characterization of immunoglobulin A1 proteases from Bacteroides and Capnocytophaga spp. Infect. Immun. 55:631–638.,31Frandsen, E. V. G., and W. G. Wade. 1996. Differentiation of human Capnocytophaga species by multilocus enzyme electrophoretic analysis and serotyping of immunoglobulin A1 proteases. Microbiology 142:441–448.,32Mortensen, S. B., and M. Kilian. 1984. Purification and characterization of an immunoglobulin A1 protease from Bacteroides melaninogenicus. Infect. Immun. 45:550–557.,33Kilian M. Degradation of immunoglobulins A2, A2, and G by suspected principal periodontal pathogens. Infect Immun. 1981 Dec;34(3):757-65.,34Frandsen, E. V. G., J. Reinholdt, and M. Kilian. 1987. Enzymatic and antigenic characterization of immunoglobulin A1 proteases from Bacteroides and Capnocytophaga spp. Infect. Immun. 55:631–638.,35Frandsen, E. V. G., and W. G. Wade. 1996. Differentiation of human Capnocytophaga species by multilocus enzyme electrophoretic analysis and serotyping of immunoglobulin A1 proteases. Microbiology 142:441–448.
- Variations of immune suppression (researchers claim this is beneficial anti-inflammatory effect of pathogenic bacteria.),36Stables MJ, Shah S, Camon EB, Lovering RC, Newson J, Bystrom J, et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood (2011) 118(26):e192–208. doi:10.1182/blood-2011-04-345330,37Ortega-Gomez A, Perretti M, Soehnlein O. Resolution of inflammation: an integrated view. EMBO Mol Med (2013) 5(5):661–74. doi:10.1002/ emmm.201202382,38Sugimoto Michelle A., Sousa Lirlândia P., Pinho Vanessa, Perretti Mauro, Teixeira Mauro M. Resolution of Inflammation: What Controls Its Onset? Frontiers in Immunology. Vol. 7. 2016. 160
- Molecular mimicry – mimicking microbial competitors,39Christen, U., D. Benke, T. Wolfe, E. Rodrigo, A. Rhode, A. C. Hughes, M. B. Oldstone, and M. G. Von Herrath. 2004. Cure of prediabetic mice by viral infections involves lymphocyte recruitment along an IP-10 gradient. J. Clin. Invest. 113: 74–84.,40Zaccone, P., Z. Fehervari, F. M. Jones, S. Sidobre, M. Kronenberg, D. W. Dunne, and A. Cooke. 2003. Schistosoma mansoni antigens modulate the activity of the innate immune response and prevent onset of type 1 diabetes. Eur. J. Immunol. 33: 1439–1449.
- Microbes can counteract this antimicrobial function by inhibiting the expression of the inducible nitric oxide synthase (iNOS).41Nathan C. Role of iNOS in human host defense. Science. 2006; 312:1874–1875. author reply 1874–1875. [PubMed: 16809512],42Wang M, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010; 3:ra11. [PubMed: 20159852] Releasing nitric oxide to make the immune cells think the area the microbes colonize has already been cleared, produced antibodies for the microbial competitors. Microbes elicit host NO synthesis to diminish the effectiveness of Antibiotic therapy and IFN-γ-primed macrophages.43Gosselin, D., et al. 1995. Role of tumor necrosis factor alpha in innate resistance to mouse pulmonary infection with Pseudomonas aeruginosa. Infect. Immun. 63:3272–3278.,44MacMicking, J. D., et al. 1997. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 94:5243– 5248.,45Mahoney, E., et al. 2002. Bacterial colonization and the expression of inducible nitric oxide synthase in murine wounds. Am. J. Pathol. 161:2143–2152.,46Mastroeni, P., et al. 2000. Antimicrobial actions of the NADPH Phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237–247.,47Richardson, A. R., P. M. Dunman, and F. C. Fang. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61:927–939.,48Sasaki, S., et al. 1998. Protective role of nitric oxide in Staphylococcus aureus infection in mice. Infect. Immun. 66:1017–1022.,49BD. McCollister, M. Hoffman, M. Husain, A. Vázquez-Torres. Nitric Oxide Protects Bacteria from Aminoglycosides by Blocking the Energy-Dependent Phases of Drug Uptake. Antimicrobial Agents and Chemotherapy, May 2011, p. 2189–2196 Vol. 55, No. 5,50Carlos Molina-Santiago, John R. Pearson, et. al. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat Commun. 2019; 10: 1919.
Implications for Immune Function and Testing
The cleavage of immunoglobulins by microbes has significant implications for both immune function and diagnostic testing. In individuals with compromised immune systems, microbial cleavage of antibodies can exacerbate susceptibility to infections and impair the body’s ability to mount an effective immune response.

Challenges in Diagnostic Testing
Furthermore, the presence of microbial enzymes that cleave immunoglobulins can pose challenges in diagnostic testing, particularly in assays reliant on the detection of intact antibodies. The cleavage of antibodies by microbes may lead to false-negative results or inaccuracies in assessing immune status, necessitating the development of alternative testing methodologies.
Conclusion: Unraveling Microbial Strategies for Survival
In the intricate interplay between microbes and the human immune system, the phenomenon of microbial cleavage of antibodies represents a compelling survival strategy employed by various pathogens. By cleaving immunoglobulins, microbes can subvert the host’s immune defenses, establishing a foothold within the body and perpetuating infection.
Understanding these microbial survival strategies is paramount in elucidating the mechanisms underlying infectious diseases and developing novel therapeutic interventions. By unraveling the intricate tactics employed by microbes, we can strive towards bolstering immune resilience and combating microbial threats more effectively.
References
- 1Qing Han, Elizabeth M. Bradshaw, Björn Nilsson, David A. Hafler, J. Christopher Love. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab on a Chip, 2010; 10 (11): 1391 DOI: 10.1039/b926849a
- 2Koteswara R. Chintalacharuvu, Philip D. Chuang, Ashley Dragoman, et al. Cleavage of the Human Immunoglobulin A1 (IgA1) Hinge Region by IgA1 Proteases Requires Structures in the Fc region of IgA. Infect Immun. 2003 May; 71(5): 2563–2570. doi: 10.1128/IAI.71.5.2563-2570.2003 PMCID: PMC153282
- 3Fujiyama, Y., M. Iwaki, K. Hodohara, S. Hosoda, and K. Kobayashi. 1986. The site of cleavage in human alpha chains of IgA1 and IgA2: A2m(1) allotype paraprotein by the clostridial IgA protease. Mol. Immunol. 23:147–150.
- 4Fujiyama, Y., K. Kobayashi, S. Senda, Y. Benno, T. Bamba, and S. Hosoda. 1985. A novel IgA protease from Clostridium sp. capable of cleaving IgA1 and IgA2m(1) but not IgA2m(2) allotype paraproteins. J. Immunol. 134: 573–576.
- 5Anna Eriksson and Mari Norgren. Cleavage of Antigen-Bound Immunoglobulin G by SpeB Contributes to Streptococcal Persistence in Opsonizing Blood. Infect. Immun. 2003, 71(1):211. DOI: 10.1128/IAI.71.1.211-217.2003.
- 6Puthia MK, Vaithilingam A, Lu J, Tan KS.Degradation of human secretory immunoglobulin A by Blastocystis.Parasitol Res. 2005 Nov;97(5):386-9. Epub 2005 Sep 7.
- 7Lei B, DeLeo FR, Hoe NP et al. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat Med 2001; 7: 1298–1305.
- 8Garcia-Nieto RM, Rico-Mata R, Arias-Negrete S, Avila EE. Degradation of human secretory IgA1 and IgA2 by Entamoeba histolytica surface-associated proteolytic activity. Parasitol Int. 2008 Dec;57(4):417-23. Epub 2008 May 15.
- 9Xuchu Que and Sharon L. Reed. Cysteine Proteinases and the Pathogenesis of Amebiasis Clin Microbiol Rev. 2000 April; 13(2): 196–206. PMCID: PMC100150
- 10E V Frandsen, J Reinholdt, and M Kilian. Enzymatic and antigenic characterization of immunoglobulin A1 proteases from Bacteroides and Capnocytophaga spp. Infect Immun. 1987 March; 55(3): 631–638. PMCID: PMC260386
- 11Gregory RL, Kim DE, Kindle JC, Hobbs LC, Lloyd DR.Immunoglobulin-degrading enzymes in localized juvenile periodontitis. J Periodontal Res. 1992 May;27(3):176-83.
- 12Senior BW, Dunlop JI, Batten MR, Kilian M, Woof JM. Cleavage of a recombinant human immunoglobulin A2 (IgA2)-IgA1 hybrid antibody by certain bacterial IgA1 proteases. Infect Immun. 2000 Feb;68(2):463-9.
- 13Frandsen EV, Reinholdt J, Kjeldsen M, Kilian M. In vivo cleavage of immunoglobulin A1 by immunoglobulin A1 proteases from Prevotella and Capnocytophaga species. Oral Microbiol Immunol. 1995 Oct;10(5):291-6.
- 14Qiu J, Brackee GP, Plaut AG. Analysis of the specificity of bacterial immunoglobulin A (IgA) proteases by a comparative study of ape serum IgAs as substrates. Infect Immun. 1996 Mar;64(3):933-7.
- 15Reinholdt J, Kilian M. Interference of IgA protease with the effect of secretory IgA on adherence of oral streptococci to saliva-coated hydroxyapatite. J Dent Res. 1987 Feb;66(2):492-7.
- 16Cizelj I, Bercic RL, Dusanic D, Narat M, Kos J, Dovc P, Bencina D. Mycoplasma gallisepticum and Mycoplasma synoviae express a cysteine protease CysP, which can cleave chicken IgG into Fab and Fc. Microbiology. 2011 Feb;157(Pt 2):362-72. Epub 2010 Oct 21.
- 17Morse SA. Neisseria, Moraxella, Kingella and Eikenella. In: Baron S, editor. SourceMedical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 14.
- 18Senior BW, Batten MR, Kilian M, Woof JM. Amino acid sequence requirements in the human IgA1 hinge for cleavage by streptococcal IgA1 proteases. Biochem Soc Trans. 2002 Aug;30(4):516-8.
- 19Hulting G, Flock M, Frykberg L, Lannergård J, Flock JI, Guss B. Two novel IgG endopeptidases of Streptococcus equi. FEMS Microbiol Lett. 2009 Sep;298(1):44-50.
- 20Kharazmi A. Mechanisms involved in the evasion of the host defence by Pseudomonas aeruginosa. Immunol Lett. 1991 Oct;30(2):201-5.
- 21Silva BA, Pinto MR, Soares RM, Barreto-Bergter E, Santos AL. Pseudallescheria boydii releases metallopeptidases capable of cleaving several proteinaceous compounds. Res Microbiol. 2006 Jun;157(5):425-32. Epub 2006 Jan 20.
- 22Loukas A. Prociv. Immune Responses in Hookworm Infections. Clin Microbiol Rev. 2001 October; 14(4): 689-703
- 23Pritchard DI. The survival strategies of hookworms. Parasitol Today. 1995;11:255-259
- 24McIntosh RS. Jones FM. Dunne DW. McKerrow JH. Pleass RJ. Characterization of immunoglobulin binding by schistosomes. Parasite Immunology 2006, 28, 407-419
- 25Faherty CS, Maurelli AT. Staying alive: bacterial inhibition of apoptosis during infection. Trends Microbiol. 2008;16(4):173-180. doi:10.1016/j.tim.2008.02.001
- 26JA. Carrero, ER. Unanue. Lymphocyte apoptosis as an immune subversion strategy of microbial pathogens. Trends in Immunology, Vol 27, ISSUE 11, P497-503, NOVEMBER 01, 2006, DOI:https://doi.org/10.1016/j.it.2006.09.005
- 27Hotchkiss RS, Tinsley KW, Swanson PE, et al. Prevention of lymphocyte cell death in sepsis improves survival in mice. Proc Natl Acad Sci U S A. 1999;96(25):14541-14546. doi:10.1073/pnas.96.25.14541
- 28Frandsen, E. V. G., J. Reinholdt, and M. Kilian. 1991. Immunoglobulin A1(IgA1) proteases from Prevotella (Bacteroides) and Capnocytophaga species in relation to periodontal diseases. J. Periodontal Res. 26:297–299.
- 29Frandsen, E. V. G., J. Reinholdt, M. Kjeldsen, and M. Kilian. 1995. In vivo cleavage of immunoglobulin A1 by immunoglobulin A1 proteases from Prevotella and Capnocytophaga species. Oral Microbiol. Immunol. 10:291–296.
- 30Frandsen, E. V. G., J. Reinholdt, and M. Kilian. 1987. Enzymatic and antigenic characterization of immunoglobulin A1 proteases from Bacteroides and Capnocytophaga spp. Infect. Immun. 55:631–638.
- 31Frandsen, E. V. G., and W. G. Wade. 1996. Differentiation of human Capnocytophaga species by multilocus enzyme electrophoretic analysis and serotyping of immunoglobulin A1 proteases. Microbiology 142:441–448.
- 32Mortensen, S. B., and M. Kilian. 1984. Purification and characterization of an immunoglobulin A1 protease from Bacteroides melaninogenicus. Infect. Immun. 45:550–557.
- 33Kilian M. Degradation of immunoglobulins A2, A2, and G by suspected principal periodontal pathogens. Infect Immun. 1981 Dec;34(3):757-65.
- 34Frandsen, E. V. G., J. Reinholdt, and M. Kilian. 1987. Enzymatic and antigenic characterization of immunoglobulin A1 proteases from Bacteroides and Capnocytophaga spp. Infect. Immun. 55:631–638.
- 35Frandsen, E. V. G., and W. G. Wade. 1996. Differentiation of human Capnocytophaga species by multilocus enzyme electrophoretic analysis and serotyping of immunoglobulin A1 proteases. Microbiology 142:441–448.
- 36Stables MJ, Shah S, Camon EB, Lovering RC, Newson J, Bystrom J, et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood (2011) 118(26):e192–208. doi:10.1182/blood-2011-04-345330
- 37Ortega-Gomez A, Perretti M, Soehnlein O. Resolution of inflammation: an integrated view. EMBO Mol Med (2013) 5(5):661–74. doi:10.1002/ emmm.201202382
- 38Sugimoto Michelle A., Sousa Lirlândia P., Pinho Vanessa, Perretti Mauro, Teixeira Mauro M. Resolution of Inflammation: What Controls Its Onset? Frontiers in Immunology. Vol. 7. 2016. 160
- 39Christen, U., D. Benke, T. Wolfe, E. Rodrigo, A. Rhode, A. C. Hughes, M. B. Oldstone, and M. G. Von Herrath. 2004. Cure of prediabetic mice by viral infections involves lymphocyte recruitment along an IP-10 gradient. J. Clin. Invest. 113: 74–84.
- 40Zaccone, P., Z. Fehervari, F. M. Jones, S. Sidobre, M. Kronenberg, D. W. Dunne, and A. Cooke. 2003. Schistosoma mansoni antigens modulate the activity of the innate immune response and prevent onset of type 1 diabetes. Eur. J. Immunol. 33: 1439–1449.
- 41Nathan C. Role of iNOS in human host defense. Science. 2006; 312:1874–1875. author reply 1874–1875. [PubMed: 16809512]
- 42Wang M, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010; 3:ra11. [PubMed: 20159852]
- 43Gosselin, D., et al. 1995. Role of tumor necrosis factor alpha in innate resistance to mouse pulmonary infection with Pseudomonas aeruginosa. Infect. Immun. 63:3272–3278.
- 44MacMicking, J. D., et al. 1997. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 94:5243– 5248.
- 45Mahoney, E., et al. 2002. Bacterial colonization and the expression of inducible nitric oxide synthase in murine wounds. Am. J. Pathol. 161:2143–2152.
- 46Mastroeni, P., et al. 2000. Antimicrobial actions of the NADPH Phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237–247.
- 47Richardson, A. R., P. M. Dunman, and F. C. Fang. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61:927–939.
- 48Sasaki, S., et al. 1998. Protective role of nitric oxide in Staphylococcus aureus infection in mice. Infect. Immun. 66:1017–1022.
- 49BD. McCollister, M. Hoffman, M. Husain, A. Vázquez-Torres. Nitric Oxide Protects Bacteria from Aminoglycosides by Blocking the Energy-Dependent Phases of Drug Uptake. Antimicrobial Agents and Chemotherapy, May 2011, p. 2189–2196 Vol. 55, No. 5
- 50Carlos Molina-Santiago, John R. Pearson, et. al. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat Commun. 2019; 10: 1919.
